Custom peptide synthesis
Custom High-Throughput Peptides and Peptide Libraries
Peptide libraries are widely used for screening purposes, allowing a quick identification of bioactive peptides at an affordable price.
Benefits of peptide libraries service
Quality
Reliable custom peptide synthesis
Assistance
Superior technical design assistance upon request
Price
Cost effective screening solution
Peptide Library types
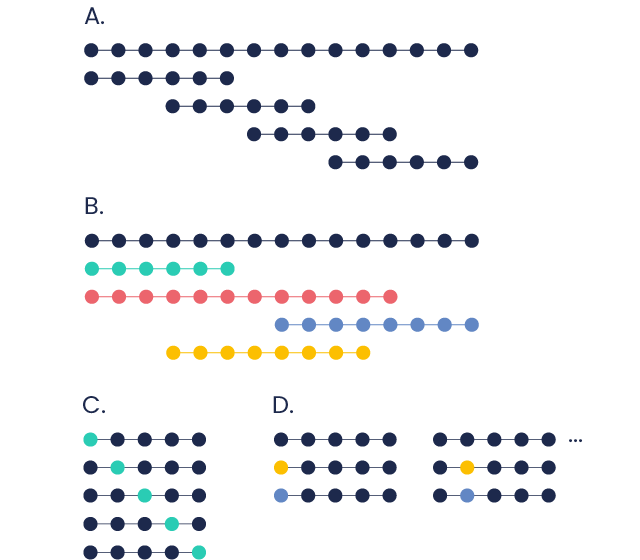
A. Libraries of overlapping peptide sequences
They are usually designed from a larger protein to quickly identify a protein region contributing to a specific functionality. By individually testing the peptides, the protein sequence is screened for the presence of key epitopes.
A well-known application is epitope mapping
Overlapping peptide libraries can also be screened for identifying the peptidic region within an antigenic protein which is responsible for a T-cell response.
B. Libraries of unrelated peptides
Peptides of different lengths, are a cost-effective solution for simultaneously screening a large set of peptides in a defined application
C. Alanine scanning libraries
Each amino acid position of the peptide is sequentially replaced with an alanine.
Alanine, the smallest chiral amino acid, is used to substitute each non-alanine residue within the sequence while not drastically affecting the conformation of that protein region.
This allows identifying the specific amino acid residues contributing to a peptide function, e.g. a protein-protein interaction or ligand-receptor binding activity.
D. Positional scanning libraries
These are designed by substituting -individually and sequentially- each amino acid with a selection of two other amino acids. This method is frequently used for sequence optimization. The resulting crude mixture is composed mainly of the two corresponding peptides, not necessarily in equimolar amount.